- Integrative multi-omics analyses
- Transcriptional and epigenetic regulation in DNA virus infections
- Promoter-proximal pausing
- Premature and disrupted transcription termination
- Standardization of data analysis
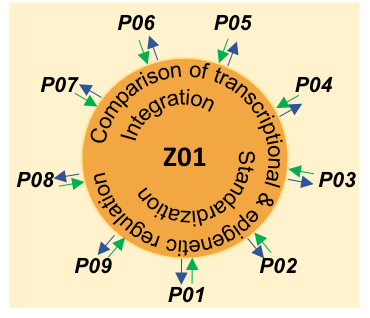
A central aim of DEEP-DV is the identification of common mechanisms and differences in host and viral gene regulation between DNA virus infections. For this purpose, we will leverage functional genomics sequencing data generated by the individual DEEP-DV projects to address their specific project objectives. These data provide a rich resource for Z01 for comparative analyses that go beyond the individual project aims and integrate across DEEP-DV projects and different functional genomics assays. To facilitate data integration, we will develop standardized analysis workflows, a data and analysis sharing platform (the DEEP-DV Hub) as well as approaches for mining public data. This will allow us to perform an extensive analysis of transcriptional and epigenetic regulation across different DNA viruses using both DEEP-DV-internal and public data. We will develop methods to investigate and compare transcriptional and epigenetic changes and their interplay during infection with different DNA viruses as well as epigenetic priming prior to infection for rapid transcriptional responses. In addition, we will focus on promoter proximal RNA Polymerase II (RNAPII) pausing, premature transcription termination and their interplay in HSV-1 and other DNA virus infections. These research questions arise from preliminary observations on a widespread RNAPII pausing loss in HSV-1 infection and premature transcription termination at polyadenylation sites. The latter has previously been associated with inhibition of the CDK9 kinase. Notably, CDK9 is required for the release of paused RNAPII from the promoter and its inhibition arrests RNAPII in a paused state. Thus, Z01 will resolve the heterogeneous roles of CDK9 during HSV-1 infection as well as the role of the HSV-1 immediate-early protein ICP22, which is known to interact with CDK9. All results will be incorporated into the DEEP-DV Hub, which will provide the foundation for a public knowledgebase on DNA virus infections pursued in a putative second funding period.
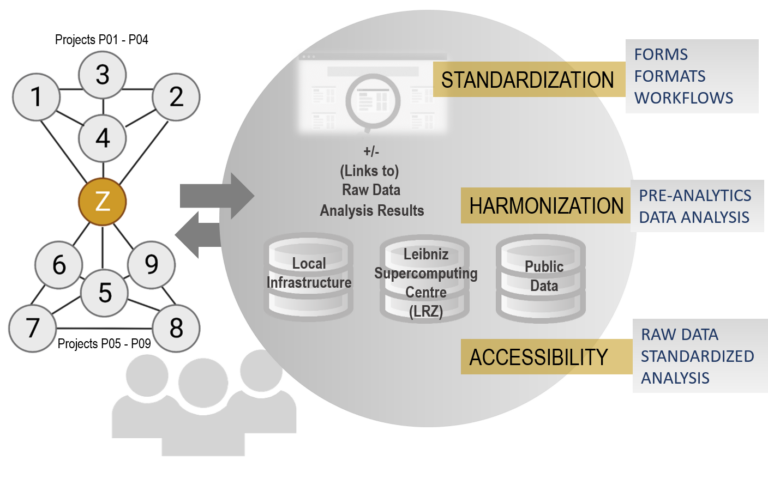
To answer these questions, we will integrate data generated with diverse Omics techniques (e.g. ChIP-Seq/mentation, scRNA-Seq, ATAC-Seq) in individual projects of DEEP-DV.
This broad spectrum of techniques is only possible thanks to the close cooperation within DEEP-DV.