P08 - Sabrina Schreiner
Spatiotemporal regulation of host/viral chromatin remodeling complexes by Human Adenovirus (HAdV) determinants during early infection
- Manipulation of host cells by human adenoviruses (HAdV)
- Role of PML nuclear bodies/SUMOylation in virus infection
- Virus-Host interplay
- Development of novel antiviral agents
- Chronic infection – elimination of the persistence reservoir
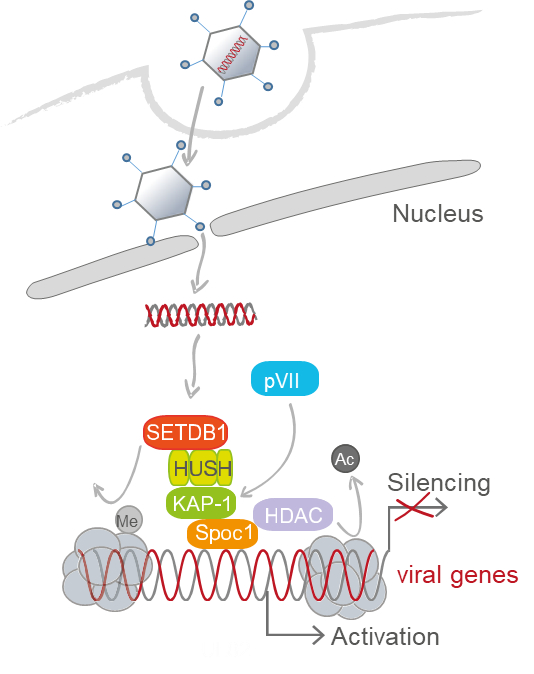
Human Adenoviruses (HAdVs) are widespread pathogens causing fatal diseases in immunocompromised patients. Similar to other human viruses, HAdV can establish persistent infections. However, the characteristics and the mechanism behind are poorly understood. Here, we will analyze HAdV-mediated manipulation of initial viral and cellular transcription conferred by SUMO-dependent PML-NBs and chromatin-associated SPOC1/KAP/HUSH complexes, prior to de novo virus gene expression. We will shed light on novel host factors/processes and SUMO PTM that allow delivery of viral genomes to specific sites in the nucleus where conversion into a transcriptional active state and selective modulation of host transcription take place. We propose that HAdV pVII is an activator of viral gene expression by antagonizing repressive complexes and orchestrating decisions into either HAdV productive infection or persistent life cycles. We will investigate if any spatiotemporal deregulation of these complexes might induce silencing of viral transcription and modulation of IFN signaling to promote a persistent infection. The central question of the project is how HAdV establish and maintain persistence by manipulating the host responses, host SUMO PTM events and immune response to silence viral transcription. By using mass spectrometry, chromatin immunoprecipitation sequencing and RNA-Seq, we will study the function and regulation of pVII in infected T-lymphocytes (HAdV-B26), epithelial intestine cells (HAdV-F41) and bronchioepithelial cells (HAdV-C5). We will get comprehensive insight into spatiotemporal control of viral transcription, IFN regulation and respective viral/host regulators. Our proposed work will shed light on unknown regulatory events during initial steps of infection that allow HAdV to undergo productive infection or to persist in human cells regulated by the specific interplay of viral and host factors.
To answer these questions we are applying multiple tools including DNA live-cell labeling technologies, RNA/ChIP-Seq analysis, and mass spectrometry analysis in appropriate infection models.
This broad spectrum of techniques is only possible thanks to the close cooperation within DEEP-DV.


