
P07 - Thomas Stamminger
Spatiotemporal control of human cytomegalovirus transcriptional silencing during lytic and latent infection
- Human Cytomegalovirus
- Silencing factors
- Repressor occupancy of viral genome
- Transcriptional signature dictating silencing
- Visualization of viral genome silencing
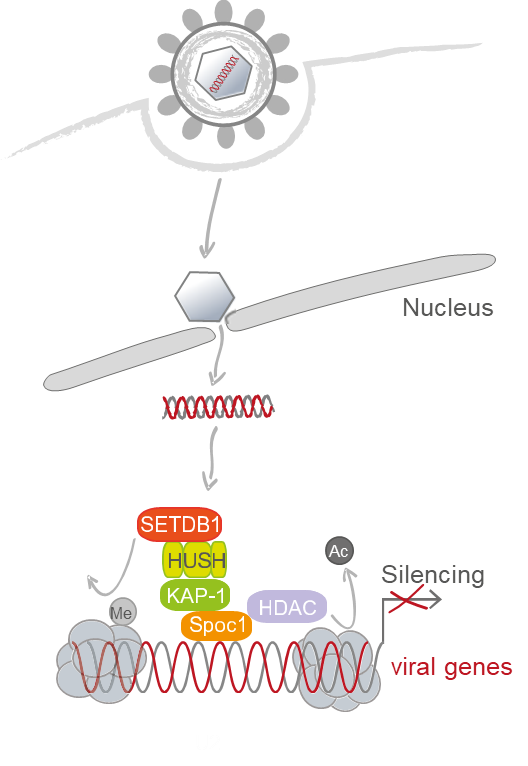
Human cytomegalovirus (HCMV), a species-specific beta-herpesviruses, persists as a subclinical, lifelong infection in the normal human host. However, under conditions of immature or compromised immune control reactivation from latency frequently causes severe disease. Previous studies demonstrated that the HCMV genome undergoes transcriptional silencing during both lytic and latent infection, which is mediated by cellular heterochromatin building factors. So far, the molecular mechanisms governing the silencing of HCMV gene expression are still incompletely defined. We have shown that a group of cellular restriction factors (PML, Sp100, DAXX, ATRX, SPOC1) associating with parental viral genomes in a spatially and/or temporally regulated manner contribute to transcriptional silencing. In particular, we recently observed that the chromatin-remodeling factor SPOC1 interacts with the HCMV major immediate early promoter correlating with a shutdown of viral gene expression. Since SPOC1 recruits KAP1 and could be detected in association with the Human Silencing Hub (HUSH) complex one major aim of this proposal will be to investigate the contribution of these factors to the control of HCMV transcriptional silencing during both permissive infection and latency. The second objective will be to analyze viral chromatin accessibility and repressor occupancy in a genome-wide manner using novel methods allowing for efficient epigenomic profiling of small samples. The obtained results should help to understand how repressor occupancy dictates viral chromatin states and, consequently, determines the switch between silencing and activation of viral gene expression. This will be complemented by single cell RNA-Seq investigating whether a specific host transcriptional signature is associated with HCMV transcriptional silencing as well as live cell imaging experiments to visualize the silencing of individual HCMV genomes. In collaboration with other projects of the RU we aim at a comparative view of how repressor occupancy dictates distinct viral chromatin states and, consequently, determines the gene expression profile in various DNA viruses.
To answer these questions we are applying multiple tools including, ATAC-Seq, chromatin immunoprecipitation sequencing (ChIP-Seq, CUT&Tag), RNA-Seq, and single cell RNA-Seq in appropriate infection models.
This broad spectrum of techniques is only possible thanks to the close cooperation within DEEP-DV.


