
P06 - Benedikt Kaufer
Role of the intercellular heterogeneity, epigenetics and viral factors in the decision between lytic replication and latency of human herpesvirus 6
- Herpesvirus latency
- Human Herpesvirus 6
- Herpesvirus genome integration into host telomeres
- Decision between lytic replication and latency
- Epigenetic modifications
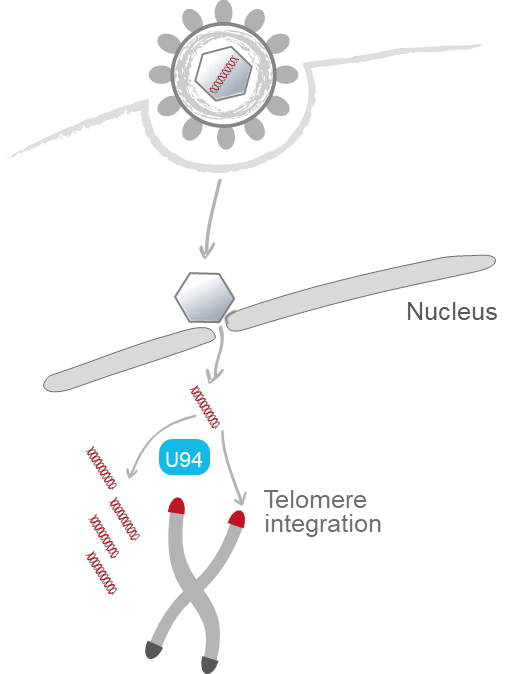
Human herpesvirus 6 (HHV-6) infects almost all humans within the first two years of life, causes Roseola infantum and is linked to a number of severe diseases. Upon primary infection, HHV-6 establishes a persistent infection in the host for life, known as latency. In latently infected cells, the HHV-6 genome can integrate into the telomeres, ensuring the maintenance of the virus genome. Intriguingly, infection of PBMCs and various clonal T-cell lines with HHV-6 can result in either lytic replication or latency. Beyond that, it remains unknown, why some cells of a clonal population support lytic replication, while the virus establishes latency in others. Importantly, individual cells of a clonal cell line differ in their transcriptional state, indicating that these differences in gene expression could influence the lytic/latent decision. In addition, we recently demonstrated that latent HHV-6 genomes are efficiently silenced upon infection using next generation sequencing. This coincided with a condensed chromatin state and repressive chromatin marks, suggesting that epigenetic silencing drives HHV-6 into latency. Therefore, we hypothesize that the transcriptional state of the cell, chromosomal integration of the virus genome, specific viral/cellular factors and/or epigenetic modifications on the virus genome influence the decision between lytic replication and latency. We will address this hypothesis in three specific objectives; (i) we will assess if transcriptional signatures influence the outcome of HHV-6A infection and will examine if selected differentially expressed genes influence the decision between lytic replication and latency, (ii) we will investigate the role of integration and the U94 protein in the establishment of latency using recombinant viruses that lack these factors and (iii) we examine if epigenetic modifications and cellular factors influence the outcome of HHV-6A infection. Our proposed work will shed light on the decision between lytic replication and latency of this ubiquitous human pathogen.
To answer these questions we are applying multiple tools including single cell RNA-seq, BAC mutagenesis, Integration assays, the CRISPR/Cas9 system to knockout genes and chromatin immunoprecipitation sequencing (ChIP-Seq) in appropriate infection models.
This broad spectrum of techniques is only possible thanks to the close cooperation within DEEP-DV.


