
P03 - Markus Landthaler
The role of RNA-binding proteins regulating transcription in HSV-1-infected cells and organoids
- Regulation of gene transcription
- Herpes Simplex Virus 1
- Protein-RNA interactions
- Role of RNA binding proteins in virus infection
- RNA binding proteins encoded by HSV-1
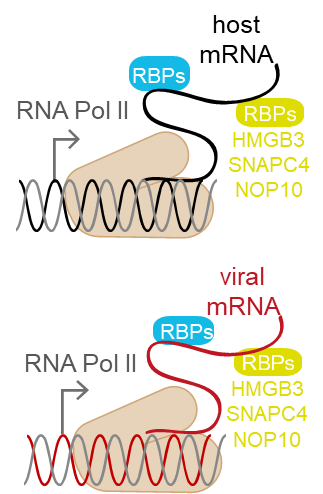
Viruses replicating in the nucleus, such as Herpesviruses, Polyomaviruses, or Adenoviruses, affect the cellular RNA metabolism in two ways. First, viral mRNA needs to be transcribed from the viral genome. Second, the cellular antiviral response leads to transcriptional and post-transcriptional changes in host gene expression. In the past decade, it became clear that RNA molecules are not merely a passive product of transcription, but themselves regulate it by many ways, for example via enhancer-associated RNAs or transcription factors-associated non-coding RNAs. All these effects are mediated by RNA-binding proteins (RBPs), which therefore play a key role in the regulation of transcription. Cellular and viral RBPs thus constitute a key layer in transcriptional regulation in virus infection. Importantly, many interferon-stimulated genes (ISGs) code for RBPs, such as the IFIT family, several RNA helicases, members of the OAS protein family, or splicing-associated proteins. Therefore, we hypothesize that RBPs are important modulators of viral transcription as well as transcription of the host’s antiviral program. Within the first funding period, we will identify host cell RBPs playing a role in HSV-1 infection, as well as RBPs coded in the viral genome by applying unbiased proteomics-based methods. The importance of the identified host cell RBPs on the progression of infection will be assessed in HEK293 cells and human cerebral organoids with loss-of-function experiments. For selected cellular and viral RBPs, target transcripts will be identified and the impact of RBPs on transcription and chromatin conformation will be examined.
To answer these questions, we are applying multiple tools based on high-throughput sequencing and data analysis, such as including mRNA interactome capture, PAR-CLIP, SLAM-Seq, 4SU-Seq, and ChIP-Seq.
This broad spectrum of techniques is only possible thanks to the close cooperation within DEEP-DV.


