
P04 - Nicole Fischer
Gene expression changes during Merkel Cell Polyomavirus productive and persistent infection in skin organoids
- Viral persistence and pathogenesis
- Human polyomaviruses (hPyV), e.g., BK polyomavirus (BKPyV) and Merkel cell polyomavirus (MCPyV)
- hPyV-induced manipulation of host gene expression
- hPyV-induced alteration of nuclear compartments
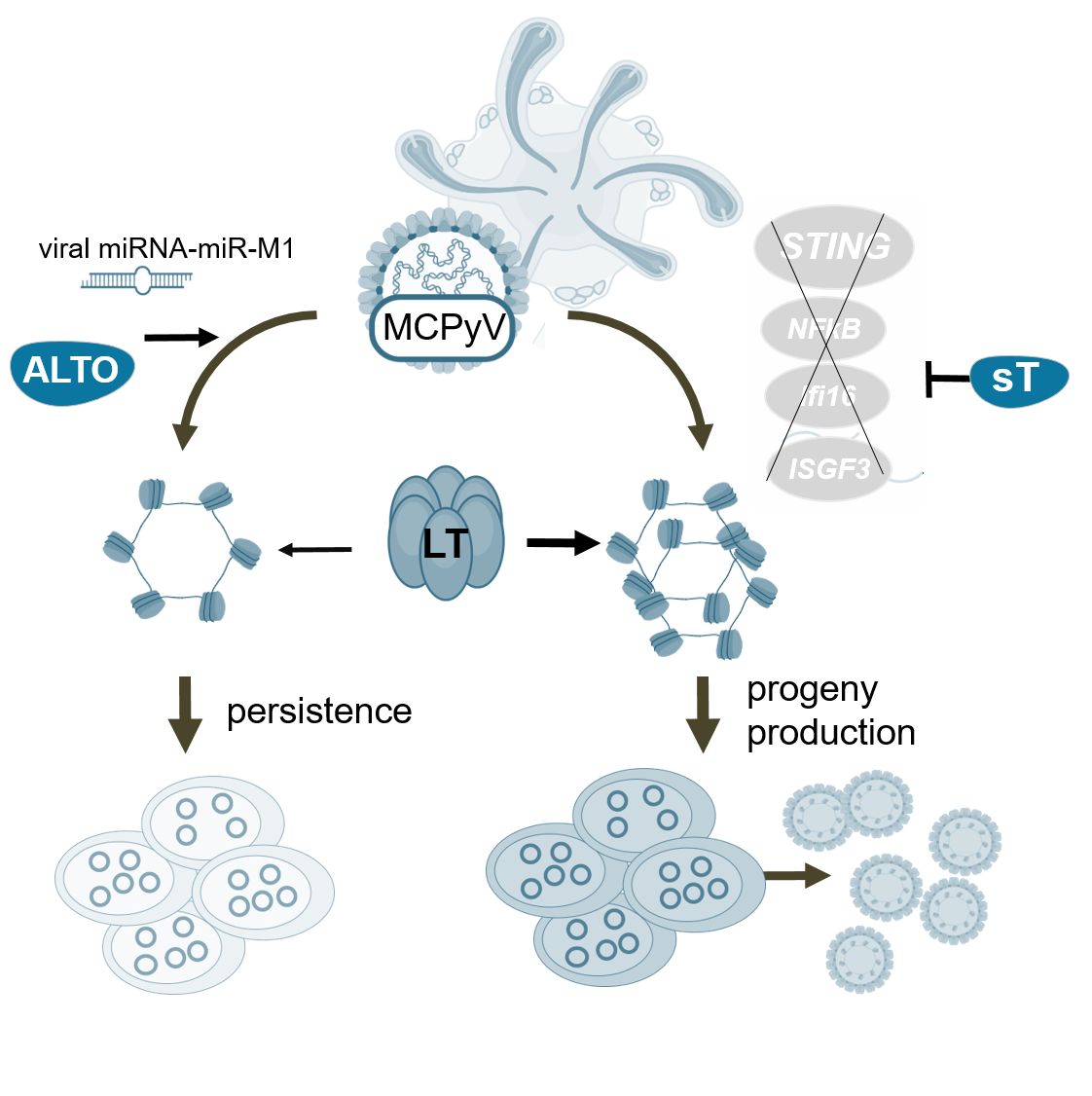
Human polyomaviruses (hPyVs) are prevalent pathogens that establish lifelong persistence in their host. These viruses are opportunistic pathogens and, under immunosuppression, contribute to uncontrolled replication and pathogenesis by reactivating persistence. A major challenge in understanding these viruses and how to combat them is the restricted cell tropism and the associated limitation in infection and pathogenicity models. Similar to many hPyVs, the cell types of persistence and the mechanisms contributing to persistence for MCPyV remain to be elucidated. The primary objectives of this project are to elucidate the molecular mechanisms that regulate productive Merkel cell polyomavirus (MCPyV) infection and to identify the factors responsible for its persistence. Building on our previously established MCPyV 3D tissue infection model, which represents the first infection model for this human tumor virus, we will analyze central molecular switches of viral infection, such as the viral proteins sT and ALTO and the host cellular components STING and IFI16, which we will target with inhibitors or genetic manipulation.
Embedded in our consortium, we will employ: single-cell RNA sequencing (scRNAseq), single-cell ATAC sequencing (scATACseq), spatial transcriptomics and advanced imaging methods available in DEEP-DV. The resulting data, as well as time-resolved data from hPyV in primary cell lines, will be incorporated into the DEEP-DV transcriptional atlas. This will facilitate the identification and understanding of factors and mechanisms of infection and persistence within polyomaviruses and across other human DNA viruses.



