Z01 - Caroline Friedel / Florian Erhard
Computational analysis of gene regulation in DNA virus infection
- Integrative multi-omics analyses
- Transcriptional and epigenetic regulation in DNA virus infections
- Promoter-proximal pausing
- Premature and disrupted transcription termination
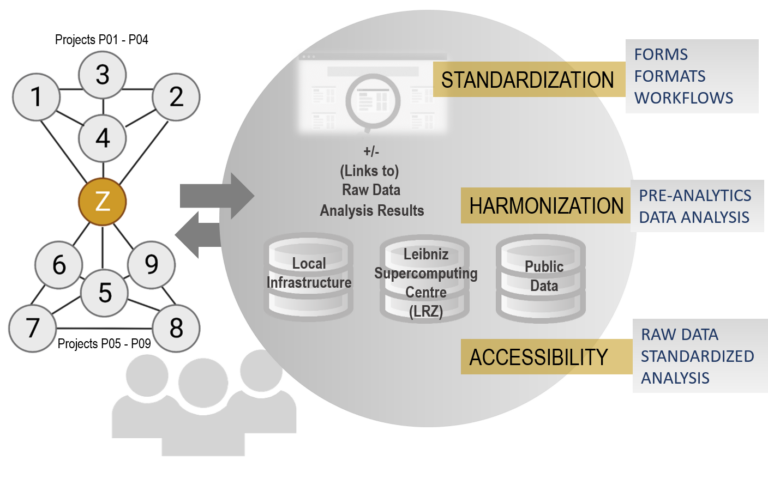
- Standardization of data analysis

In the second funding period, Z01 will develop and analyse the DEEP-DV Transcription Atlas of gene regulation in DNA virus infections at single-cell level. In collaboration with DEEP-DV partners, we will generate a comprehensive, temporally resolved, and harmonized atlas of infection by all DNA viruses studied in DEEP-DV. Due to use of metabolic RNA labelling with 4-thiouridine (4sU), the DEEP-DV Atlas will not only provide snapshots of gene expression throughout infection, but it will also allow dissecting and comparing regulatory processes with unprecedented details. The atlas will be disseminated via a web platform software that enables visualization and analysis of single-cell data from multiple projects and facilitate cross-virus comparisons.
We will also develop new computational approaches to make use of the atlas: We will implement a method to determine the stage of infection of single cells based on viral gene expression patterns. We will extend our previously developed method Heterogeneity-seq for the identification of common and distinct factors that govern infection outcomes for different viruses. Based on this, we will infer gene regulatory networks in DNA virus infections by directly making use of 4sU labelling to differentiate direct regulatory effects from mediating or confounding factors. A particular focus will be on regulation of interferon stimulated genes (ISGs) for which we will compile an up-to-date catalogue of ISGs and their regulation from public RNA- and ChIP-seq data. We will furthermore investigate and cross-compare alternative polyadenylation and aberrant transcription termination in DNA virus infections using the DEEP-DV Atlas. Finally, we will continue our investigations into HSV-1-induced changes in RNA polymerase II pausing and regulation of transposable elements in DNA virus infections together with DEEP-DV partners.