The DEEP-DV Research Unit (RU)
This DFG-funded Research Unit (RU) of molecular biologists, virologists, and bioinformaticians uses state of the art technologies to study and compare the consequences of nuclear DNA virus infections in different DNA virus families (Herpesviridae, Adenoviridae, Polyomaviridae).

The individual projects are grouped into two main research areas (Research Area 1 and Research Area 2) and are united by the Z01 project, which is dedicated to integrative functional genomics of the infectious disease models studied in DEEP-DV.
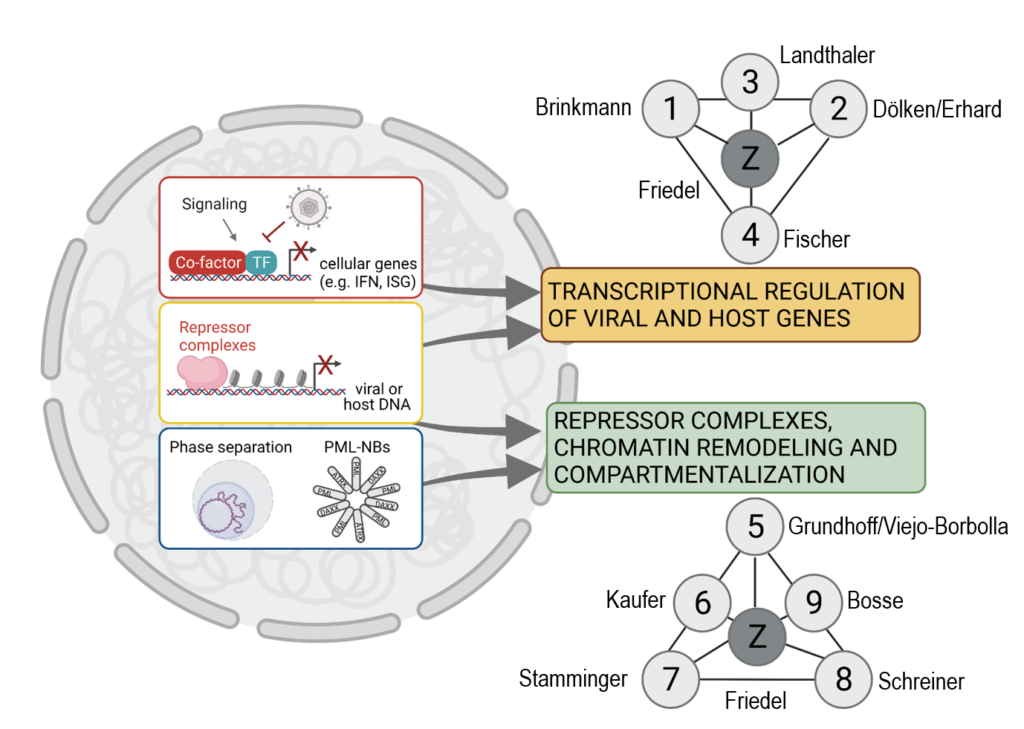
Research Area 1 (RA1) focuses on the transcriptional regulation of viral and host gene expression early after viral infection. It includes projects investigating the type I interferon (IFN) response with respect to the transcription of the type I IFN IFNB1 and interferon-stimulated genes (ISGs) downstream of pattern recognition receptor (PRR) and type I IFN receptor (IFNAR) signaling, respectively, P01 Brinkmann, P02 Dölken/Erhard, P04 Fischer, as well as research on RNA-binding proteins of viral and cellular origin that impact early transcriptional events after infection, P03 Landthaler.
Research Area 2 (RA2) focuses on repressor complexes, chromatin remodeling, and nuclear compartments. The projects synergistically address the question how viruses exploit or evade cellular chromatin regulatory pathways upon entry of a naïve viral DNA molecule into the nucleus of the infected cells. P05 Grundhoff/Viejo-Borbolla, P06 Kaufer, and P09 Bosse investigate how the cellular context and nuclear compartments can (1) promote lytic replication, or (2) lead to the acquisition of global chromatin states that either permit persistence of a transiently silenced viral genome or result in the potentially permanent inactivation of viral genomes. In addition to the role of global chromatin states, projects P07 Stamminger and P08 Schreiner investigate the role of repressor complexes recruited to specific genetic elements in the viral genome.
The expertise of the DEEP-DV members anchored in RA1 and RA2 is complemented by the bioinformatics expertise of the Friedel group (Z01), which has a strong focus on functional genomics of infectious disease models. The Friedel group has an extensive track record in integrative functional genomics data analysis and the development of new computational methods with a focus on viral infections. For example, ContextMap2 allows parallel mapping of RNA-Seq reads against multiple genomes, such as host and viral genomes, and identification of polyadenylation (pA) sites from pA-tail-containing reads. The workflow management system Watchdog supports the automated analysis of large functional genomics datasets. Friedel employs her expertise to an integrative comparison of transcriptional and epigenetic regulation in DNA virus infection (Z01), collecting data and results from all DEEP-DV projects. Additional expertise in single cell analysis and the development of bioinformatics methods specifically applicable to viral infections is contributed by the group of Florian Erhard (P02).
