
P01 - Melanie Brinkmann
Mechanistische Einblicke in die Modulation der Typ-I-Interferon-Transkription durch Tegument-Proteine des Cytomegalovirus und ihre Auswirkungen auf die virale Transkription
- Innate Immunity
- Human Cytomegalovirus
- Murine Cytomegalovirus
- Herpesviral modulation of type I interferon transcription
- Role of PML nuclear bodies for the type I IFN response
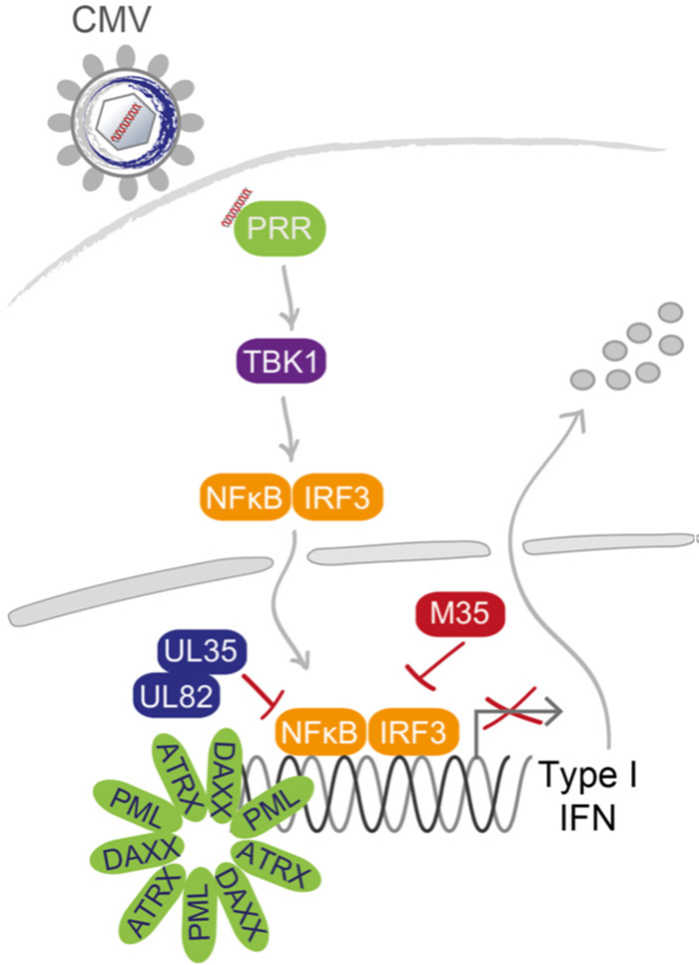
Die Erkennung einer viralen Infektion durch Mustererkennungsrezeptoren (PRR) ist entscheidend für die Einleitung einer raschen Immunantwort und die frühzeitige Kontrolle der Infektion. PRR erkennen unter anderem virale Nukleinsäuren und aktivieren daraufhin Signalkaskaden, die zur Transkription von Typ-I-Interferonen (IFN) und proinflammatorischen Zytokinen führen. Da diese antivirale Immunreaktion die virale Ausbreitung stark einschränken oder sogar verhindern würde, haben Herpesviren ihrerseits eine Reihe von eleganten Mechanismen entwickelt, die diese Immunantwort gezielt abschwächen. Herpesviren können die PRR-Signalübertragung sogar zu ihrem eigenen Vorteil nutzen, was dem fein abgestimmten Zusammenspiel zwischen Herpesviren und ihrem Wirt eine weitere Komplexitätsebene verleiht.
Virale Tegumentproteine, die mit den eindringenden Viruspartikeln in die infizierte Zelle gelangen, sind erstklassige Kandidaten für PRR-Antagonisten: Sie sind von Beginn der Infektion an in der Wirtszelle vorhanden und können so umgehend die PRR-Signalübertragung modulieren. Auf der Grundlage unserer früheren Arbeiten über herpesvirale PRR-Antagonisten werden wir in diesem Projekt die molekularen Mechanismen entschlüsseln, wie die Tegumentproteine M35, UL35 und UL82 des Cytomegalovirus (CMV) die Typ-I-IFN Genexpression im Zellkern manipulieren, und wie dies die zelluläre und virale Genexpression beeinflusst.
Da die Tegumentproteine des humanen CMV (HCMV), UL35 und UL82, in enger Nachbarschaft zu sogenannten promyelozytären Leukämie Kernkörpern (PML-NBs) lokalisieren und bereits gezeigt wurde, dass UL82 mit dem PML-NB Protein DAXX interagiert, werden wir den Beitrag verschiedener Proteinkomponenten der PML-NBs zur Typ I IFN-Transkription bei HCMV-Infektion untersuchen und diese Fragestellung gemeinsam mit anderen Mitgliedern der Forschungsgruppe bearbeiten.
Dieses Projekt wird unser Wissen über die vielfältigen Facetten der viralen Modulation der angeborenen Immunantwort vertiefen und dadurch neue Erkenntnisse über zelluläre Determinanten liefern, die für die Typ I IFN-Transkription und das Fortschreiten der DNA Virus Infektion wichtig sind.
Zur Beantwortung dieser Fragen setzen wir eine Vielzahl moderner Methoden der Molekularbiologie ein, darunter Chromatin-Immunpräzipitations-Sequenzierung (ChIP-Seq), RNA-Seq, Genom-Manipulation der Wirtszelle und Viren sowie hochentwickelte Bildgebungsanalysen.
Dieses breite Spektrum an Techniken ist nur dank der engen Zusammenarbeit innerhalb von DEEP-DV möglich.


