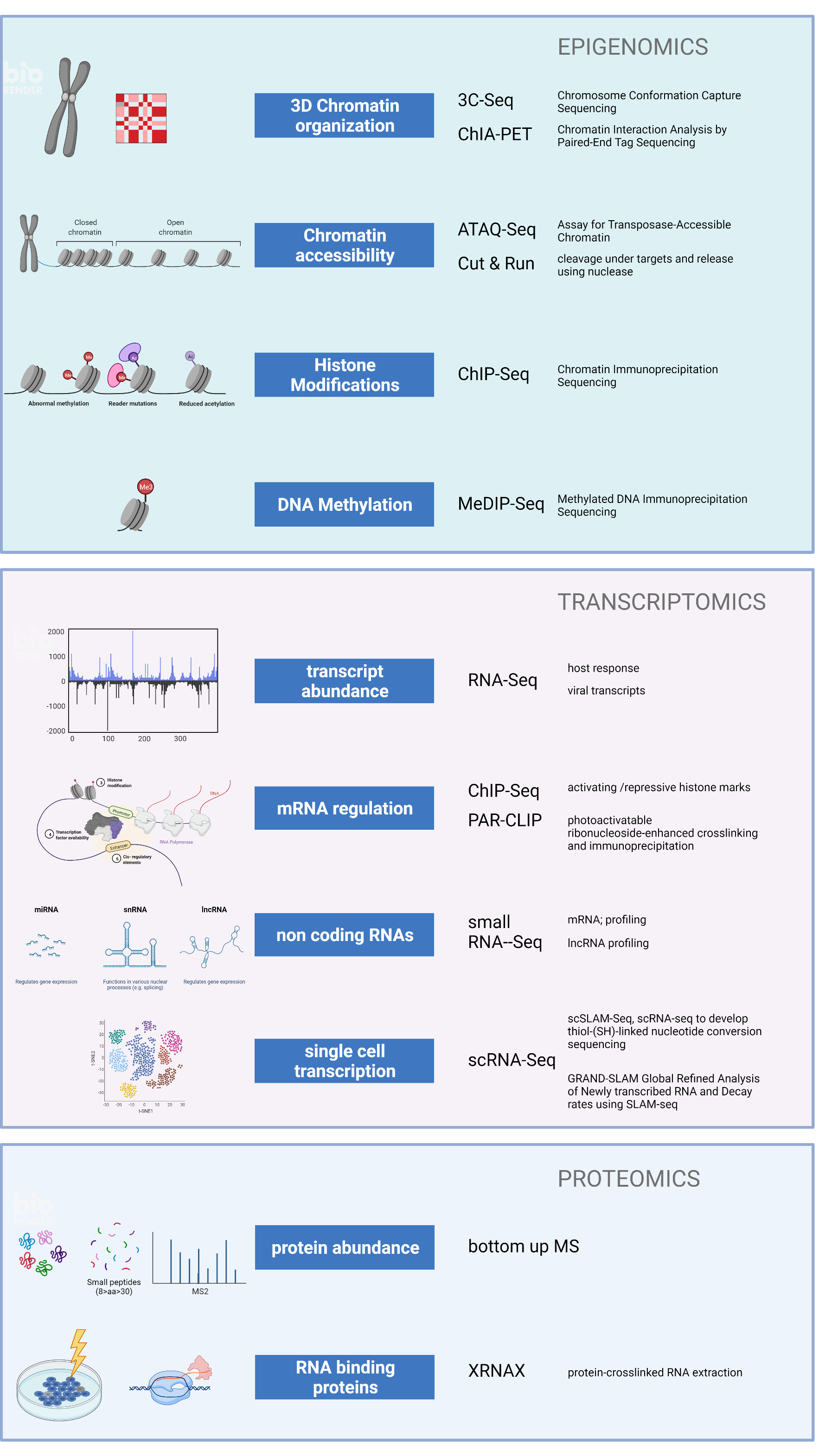
Omics
All projects in DEEP-DV combine omics-scaled analyses of bulk infections with advanced imaging or scRNA-Seq technologies to investigate heterogeneities within cell populations.
Projects in DEEP-DV apply state of the art methodologies such as omics-scale interrogation techniques (RNA-Seq, ChIP-Seq), RNA proteomics, single-cell technologies (scRNA-Seq and advanced imaging technologies), and bioinformatics to virology questions. This opens new possibilities to understand and, ultimately, control acute and chronic DNA virus infections.
To visualize genomic data and ChiPSeq data, we use publicly available software tools, e.g., igv (https://software.broadinstitute.org/software/igv/) and Easeq (https://easeq.net/).